IMC diabetes dataset, 1.8 Million cells#
This example shows some of the analysis and visualizations in SpatialTis.
It should give you a general idea on how to use it.
[1]:
%config InlineBackend.figure_format = 'retina'
import anndata as ad
import spatialtis as st
import spatialtis.plotting as sp
from spatialtis import Config
[2]:
data = ad.read_h5ad("../data/IMC-diabetes.h5ad")
data.obs.head(5)
[2]:
| area | eccentricity | islet_id | centroid | image | case | slide | part | group | stage | cell_cat | cell_type | n_genes | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 12 | 0.837664 | 0 | (52.5, 0.6666666666666666) | A01 | 6362 | A | Tail | 1 | Onset | exocrine | acinar | 30 |
| 1 | 19 | 0.902864 | 0 | (128.0, 0.894736842105263) | A01 | 6362 | A | Tail | 1 | Onset | exocrine | acinar | 36 |
| 2 | 7 | 0.882300 | 0 | (135.28571428571428, 0.42857142857142855) | A01 | 6362 | A | Tail | 1 | Onset | exocrine | acinar | 30 |
| 3 | 19 | 0.784367 | 0 | (449.5263157894737, 1.1578947368421053) | A01 | 6362 | A | Tail | 1 | Onset | exocrine | acinar | 33 |
| 4 | 30 | 0.930895 | 0 | (458.3666666666666, 1.1) | A01 | 6362 | A | Tail | 1 | Onset | exocrine | acinar | 38 |
[3]:
data
[3]:
AnnData object with n_obs × n_vars = 1776974 × 38
obs: 'area', 'eccentricity', 'islet_id', 'centroid', 'image', 'case', 'slide', 'part', 'group', 'stage', 'cell_cat', 'cell_type', 'n_genes'
var: 'markers', 'n_cells', 'highly_variable', 'means', 'dispersions', 'dispersions_norm'
uns: 'hvg'
[4]:
st.wkt_points(data, 'centroid')
[5]:
Config.exp_obs = ['stage', 'part', 'case', 'image']
Config.centroid_key = 'centroid'
Config.cell_type_key = 'cell_type'
Config.marker_key = 'markers'
Config.progress_bar = False
Check the configuration to make sure things are correct
[6]:
Config.view()
Current configurations of SpatialTis ┏━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Options ┃ Attributes ┃ Value ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ Multiprocessing │ mp │ True │ │ Verbose │ verbose │ True │ │ Progress bar │ progress_bar │ False │ │ Auto save │ auto_save │ False │ │ Experiment observations │ exp_obs │ ['stage', 'part', 'case', 'image'] │ │ ROI key │ roi_key │ image │ │ Cell type key │ cell_type_key │ cell_type │ │ Marker key │ marker_key │ markers │ │ Centroid key │ centroid_key │ centroid │ │ Shape key │ shape_key │ │ └─────────────────────────┴───────────────┴────────────────────────────────────┘
Save the config to the data
[7]:
Config.dumps(data)
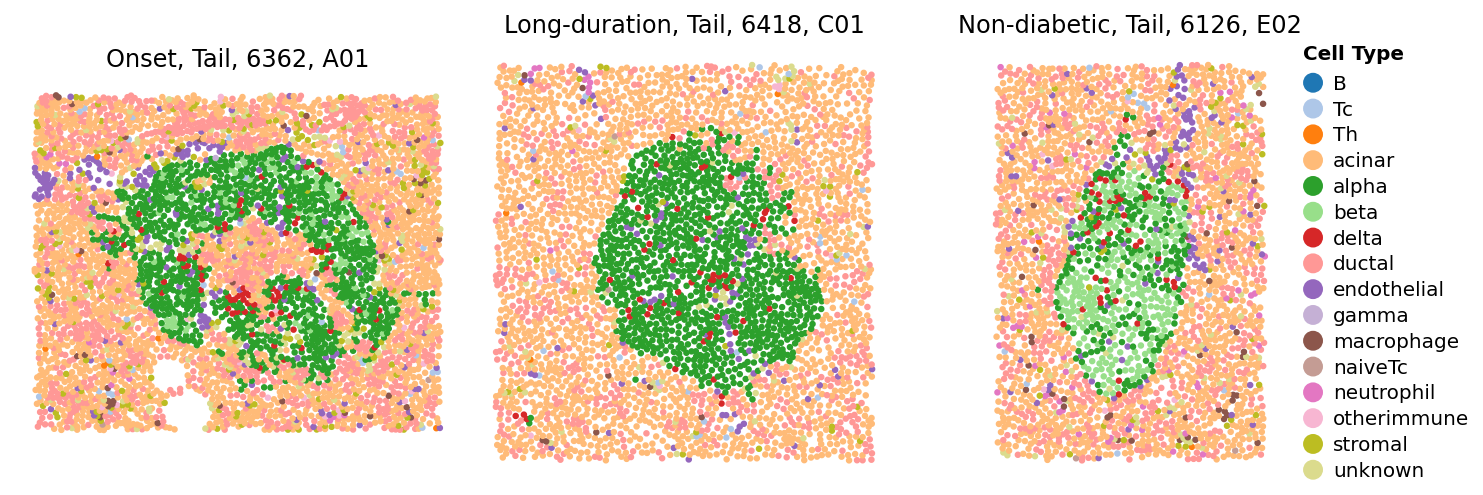
Let’s look at what those ROI looks like
[8]:
sp.cell_map(data, ['E02', 'A01', 'C01'])
[8]:
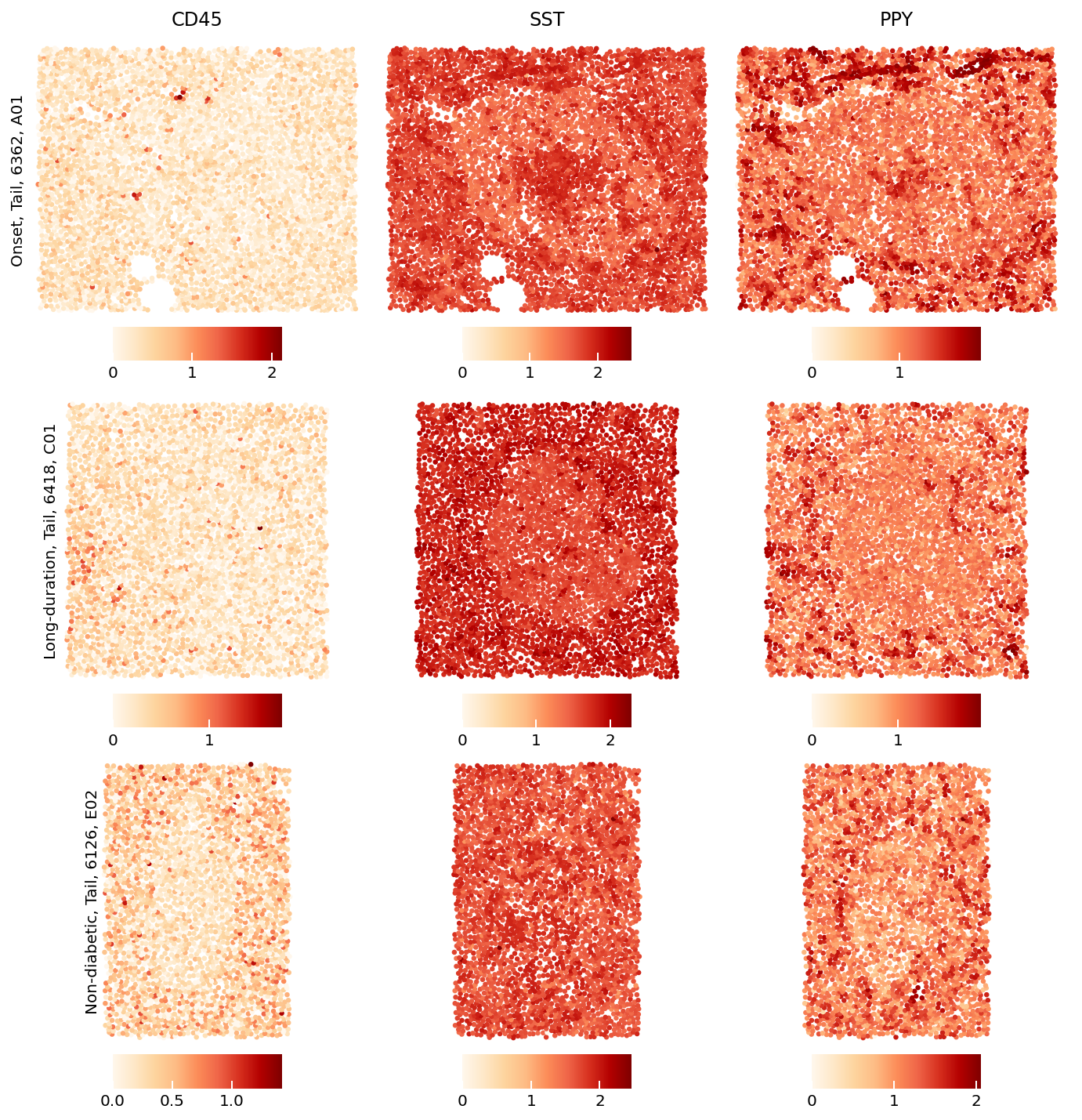
[9]:
sp.expression_map(data,
rois=['E02', 'A01', 'C01'],
markers=['CD45', 'SST', 'PPY'])
[9]:
[10]:
st.cell_components(data)
⏳ Cell components
📦 Added to AnnData, uns: 'cell_components'
⏱ 3s510ms
[11]:
sp.cell_components(data,
groupby='stage',
percentage=True,
order=['Non-diabetic', 'Onset', 'Long-duration'])
[11]:
<AxesSubplot:xlabel='stage', ylabel='Count'>
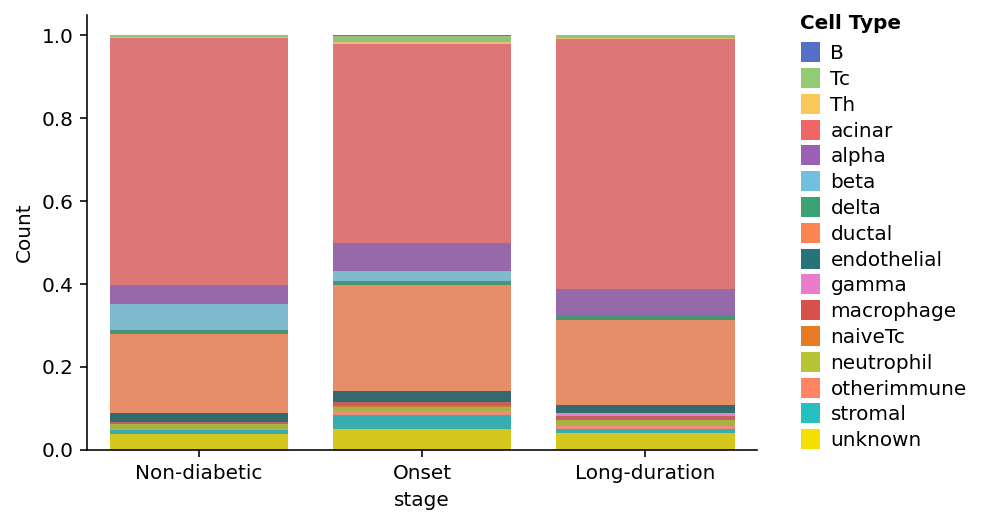
[12]:
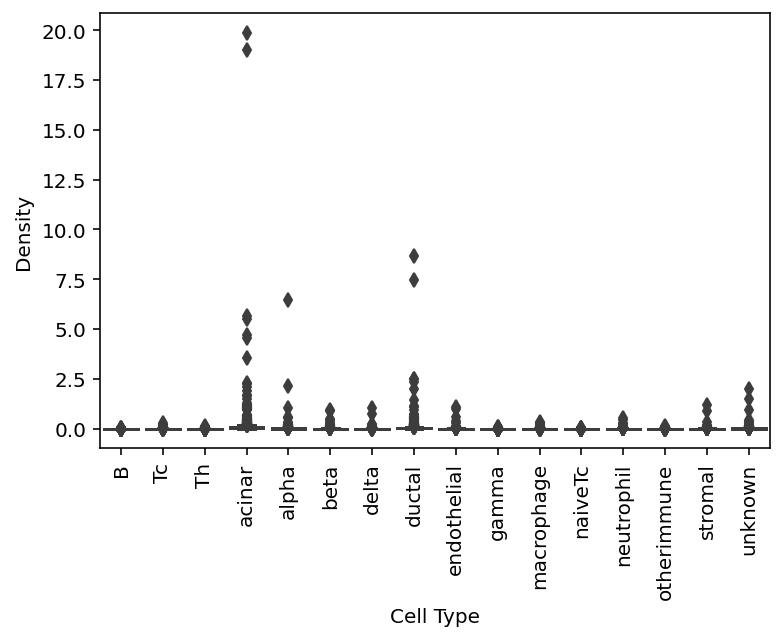
st.cell_density(data)
⏳ Cell density
📦 Added to AnnData, uns: 'cell_density'
⏱ 9s413ms
[13]:
sp.cell_density(data)
[13]:
<AxesSubplot:xlabel='Cell Type', ylabel='Density'>
[14]:
st.cell_co_occurrence(data)
⏳ Cell co-occurrence
📦 Added to AnnData, uns: 'cell_co_occurrence'
⏱ 3s455ms
[15]:
sp.cell_co_occurrence(data)
[15]:
<milkviz._dot_matrix.DotHeatmap at 0x25e129cbdf0>
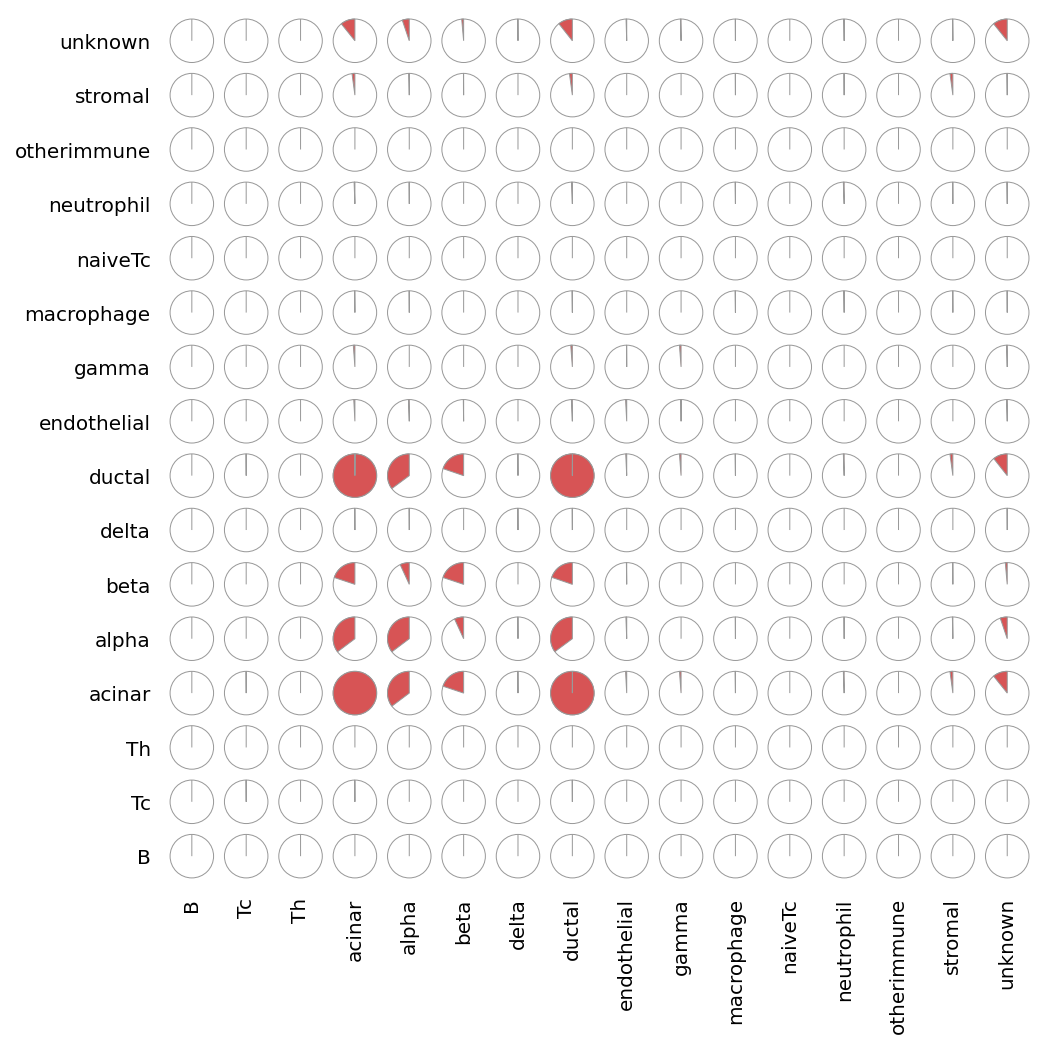
[16]:
st.cell_dispersion(data)
⏳ Cell dispersion
🛠 Method: Index of dispersion
📦 Added to AnnData, uns: 'cell_dispersion'
⏱ 9s716ms
[17]:
sp.cell_dispersion(data)
[17]:
<milkviz._dot_matrix.DotHeatmap at 0x25e8b5ce250>
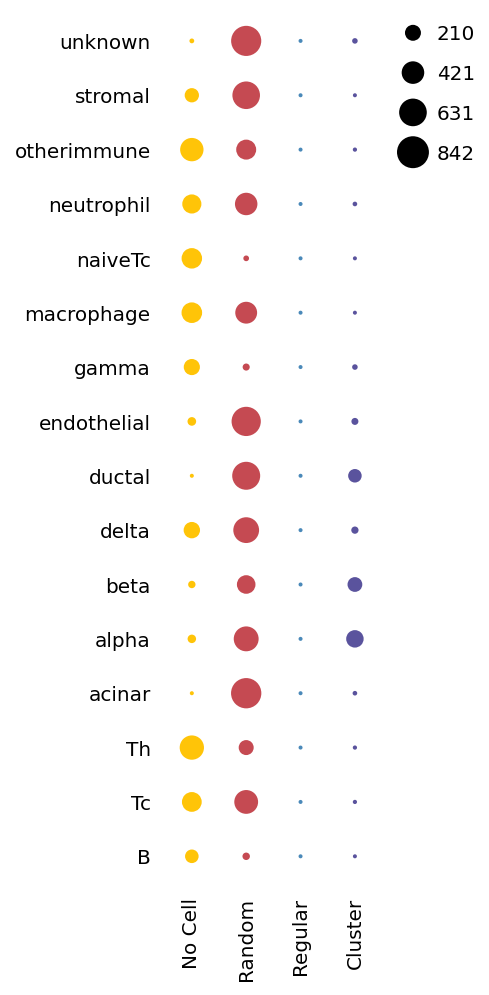
[18]:
st.spatial_heterogeneity(data)
⏳ Spatial heterogeneity
🛠 Method: Leibovici entropy
📦 Added to AnnData, uns: 'heterogeneity'
⏱ 7s587ms
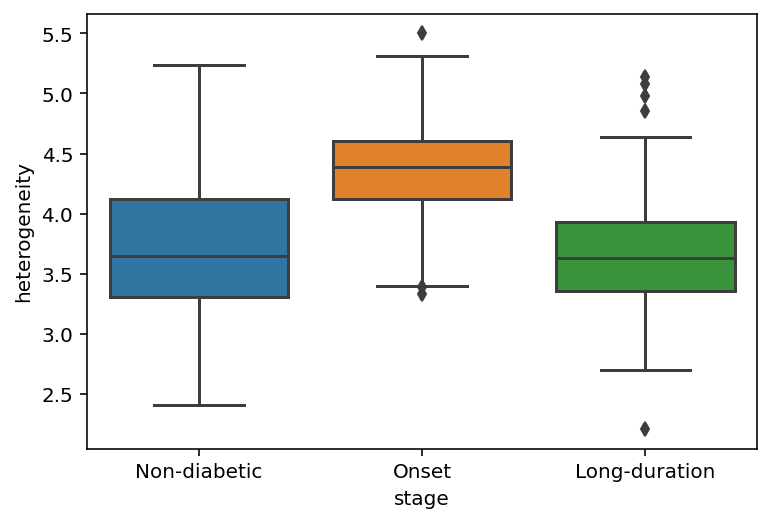
[19]:
sp.spatial_heterogeneity(data, order=['Non-diabetic', 'Onset', 'Long-duration'])
[19]:
<AxesSubplot:xlabel='stage', ylabel='heterogeneity'>
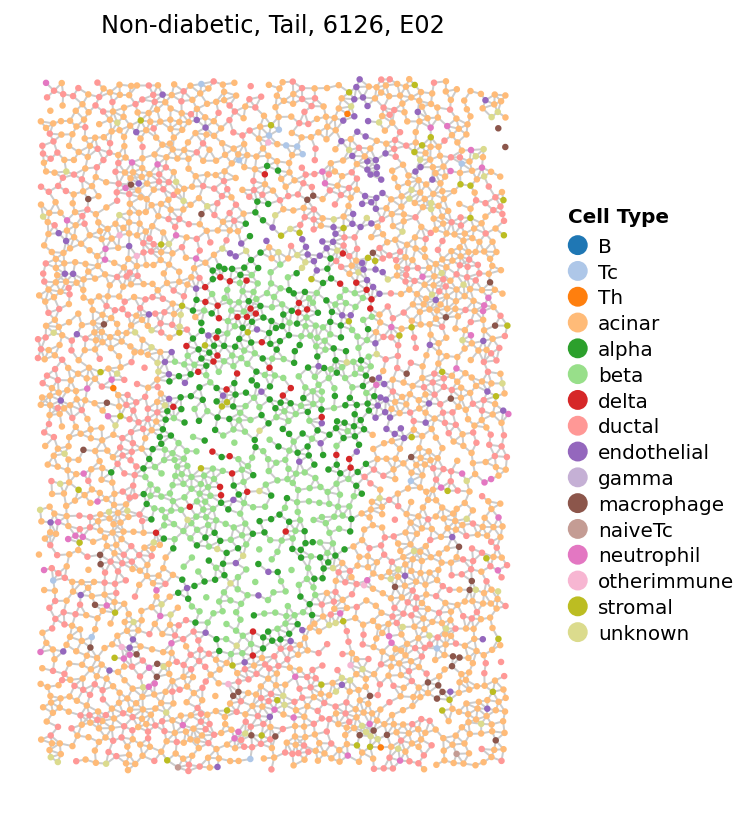
One of the most essetial steps in analyzing spatial omics data is to embedding it into a spatial network.
[20]:
st.find_neighbors(data, r=12)
⏳ Find neighbors
🛠 Method: kdtree
📦 Added to AnnData, obsm: 'cell_neighbors'
📦 Added to AnnData, obs: 'cell_neighbors_count'
⏱ 10s976ms
[21]:
sp.cell_map(data, ['E02'], show_neighbors=True, figsize=(6, 7))
[21]:
[22]:
st.cell_interaction(data)
⏳ Cell interaction
🛠 Method: pseudo p-value
📦 Added to AnnData, uns: 'cell_interaction'
⏱ 1m56s
[23]:
sp.cell_interaction(data)
[23]:
<milkviz._dot_matrix.DotHeatmap at 0x25dd0175250>
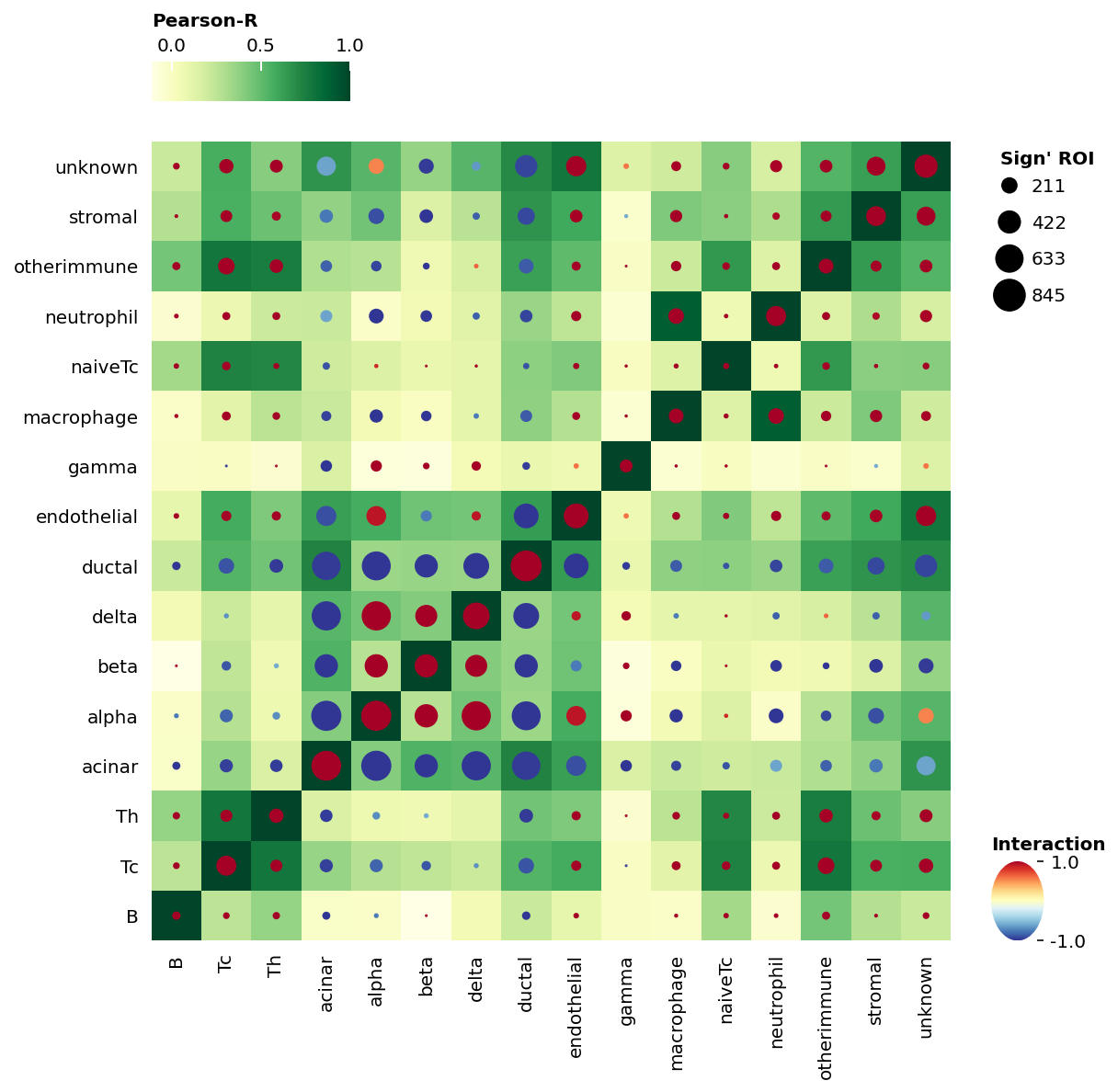
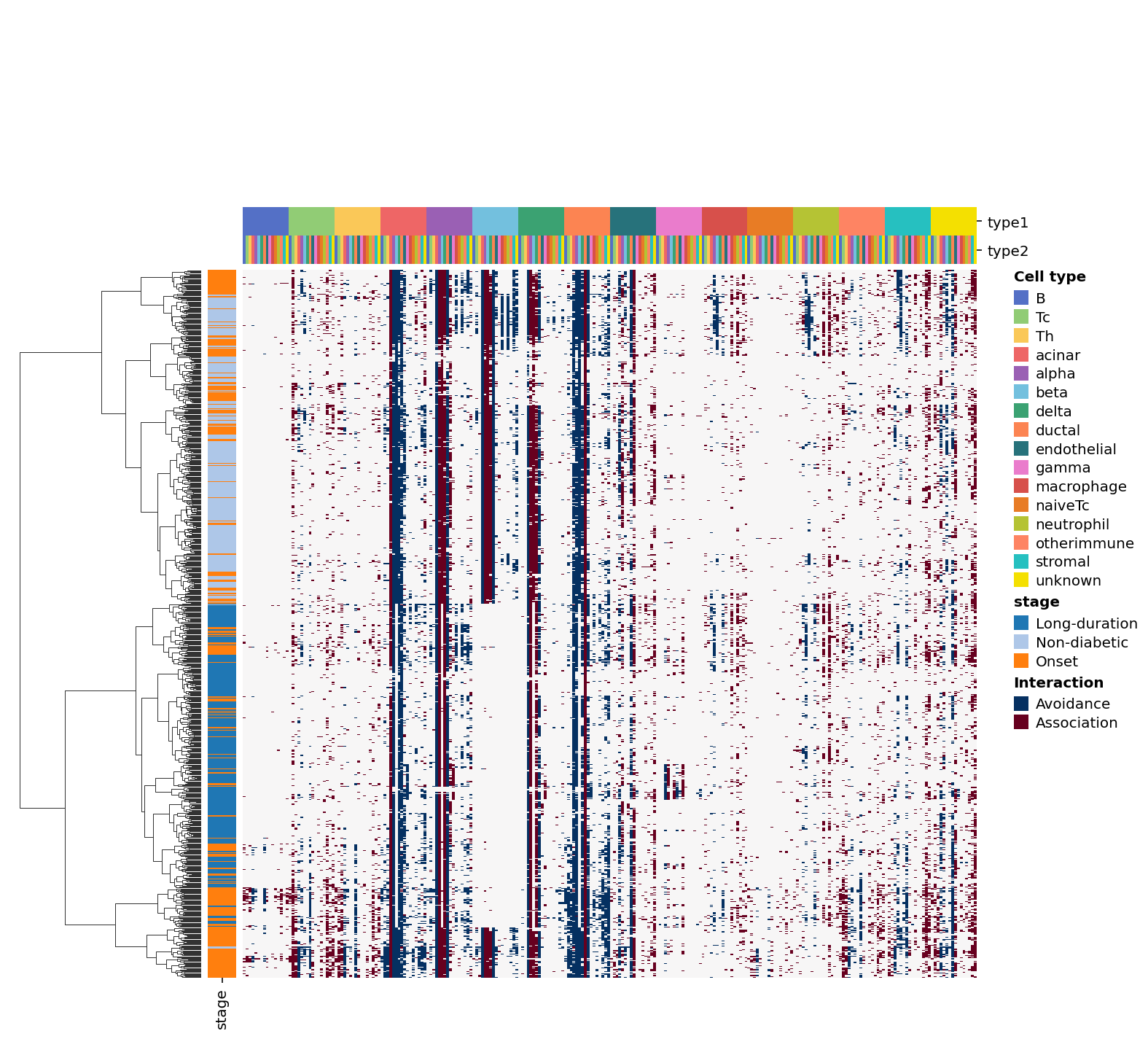
Or you can visualiza it in heatmap
[24]:
sp.cell_interaction(data, use="heatmap", groupby="stage")
[24]:
<seaborn.matrix.ClusterGrid at 0x25e449953d0>
[25]:
st.NCD_marker(data)
⏳ NCD Markers
📦 Added to AnnData, uns: 'ncd_marker'
⏱ 3m18s
[26]:
r = st.get_result(data, 'ncd_marker')
r.sort_values('log2_FC', ascending=False)
[26]:
| cell_type | marker | neighbor_type | dependency | log2_FC | pval | |
|---|---|---|---|---|---|---|
| 39 | delta | CDH | gamma | 0.914396 | 2.484879 | 3.493616e-279 |
| 34 | beta | CDH | gamma | 0.860546 | 2.417639 | 5.402380e-112 |
| 81 | unknown | CDH | gamma | 0.730400 | 2.405135 | 1.074318e-214 |
| 56 | endothelial | CDH | gamma | 0.542463 | 2.367553 | 5.895981e-90 |
| 75 | stromal | CDH | gamma | 0.528550 | 2.337943 | 2.023402e-33 |
| ... | ... | ... | ... | ... | ... | ... |
| 31 | alpha | cPARP1 | beta | 0.675172 | -0.061171 | 0.000000e+00 |
| 44 | delta | SST | beta | 0.705842 | -0.067218 | 0.000000e+00 |
| 19 | alpha | CA9 | beta | 0.673017 | -0.109999 | 0.000000e+00 |
| 37 | delta | CA9 | beta | 0.653708 | -0.142080 | 0.000000e+00 |
| 46 | delta | cCASP3 | beta | 0.516018 | -0.164905 | 6.121436e-302 |
87 rows × 6 columns
[ ]: