Spatial Analysis¶
SpatialTis provides a series of Spatial Anlaysis methods:
Find cell neighbors
Neighbor dependent markers
Cell type based analysis:
Spatial Distribution*
Spatial Heterogenenity*
Hotspot Detection*
Neighborhood Analyisis (Cell-Cell Interaction)
* No need to compute neighbors information
Markers based analysis:
Spatial Enrichment Analysis (Markers spatial enrichemnt)
Spatial Co-expression
Spatial community detection
[1]:
import warnings
warnings.filterwarnings("ignore", category=FutureWarning)
[2]:
import anndata as ad
data = ad.read_h5ad("../data/imc_data.h5ad")
data
[2]:
AnnData object with n_obs × n_vars = 1776974 × 38
obs: 'area', 'eccentricity', 'islet_id', 'centroid', 'image', 'case', 'slide', 'part', 'group', 'stage', 'cell_cat', 'cell_type'
var: 'markers'
This is a huge dataset, almost 1.8 millions of cell
[3]:
import spatialtis as st
import spatialtis.plotting as sp
from spatialtis import CONFIG
CONFIG.EXP_OBS = ["stage", "case", "part", "image"]
CONFIG.CELL_TYPE_KEY = "cell_type"
CONFIG.MARKER_KEY = "markers"
CONFIG.CENTROID_KEY = "centroid"
Since this is a diabetic data, we mainly focus on islets cells. If you are interesated in the immune part, feel free to look into it yourself.
[4]:
islets_cells = ['gamma', 'delta', 'alpha', 'beta']
group_order={"stage":["Non-diabetic", "Onset", "Long-duration"]}
Spatial Distribution¶
There are three type of distribution pattern (0 if no cells)
Random (1)
Regular (2)
Cluster (3)
Three methods are provided
Variance-to-mean ratio (vmr): Index of Dispersion
Quadratic statistics (quad): Morisita’s index of dispersion
Nearest neighbors search (nns): Clark and Evans aggregation index
Two visualizations:
Dot plot
Heatmap
[5]:
st.spatial_distribution(data)
⏳ Spatial Distribution
🛠 Method: Nearest neighbors search
Finding distribution pattern ██████████ 100% 00:00|00:13
📦 Added to AnnData, uns: 'spatial_distribution'
⏱ 58s905ms
[5]:
[6]:
sp.spatial_distribution(data)
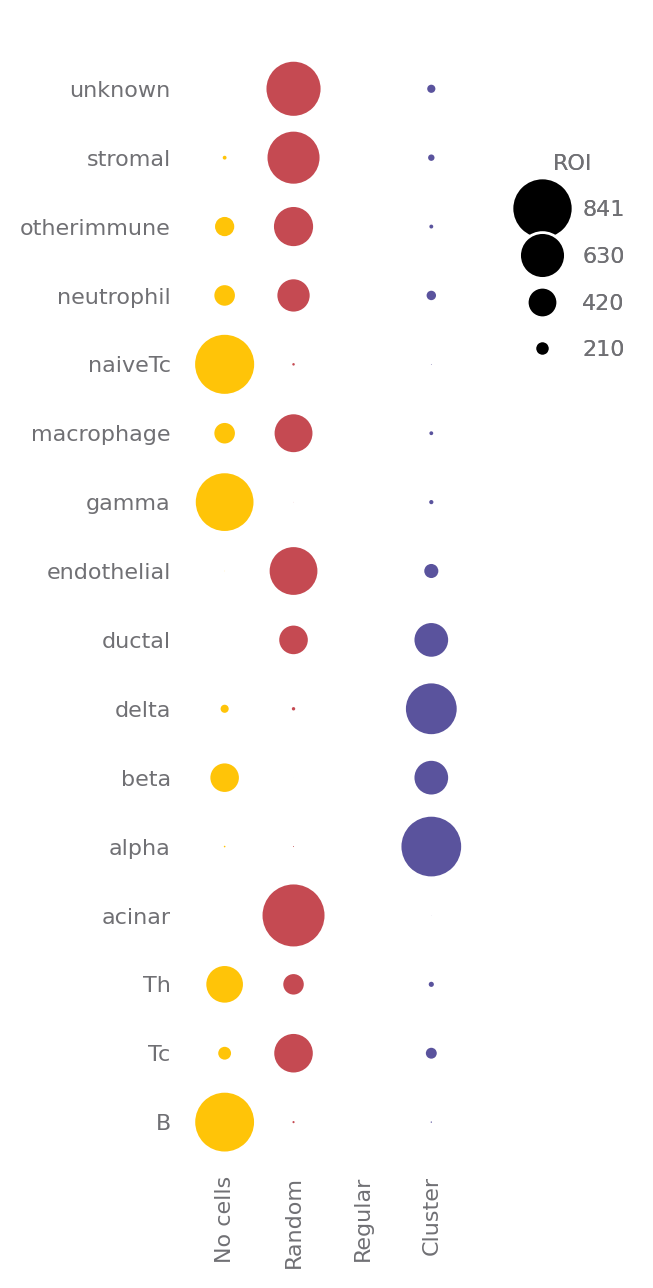
[6]:
Or you could visualize it in heatmap
[7]:
sp.spatial_distribution(data, groupby=['stage'], use="heatmap", clustermap_kwargs=dict(row_cluster=True))
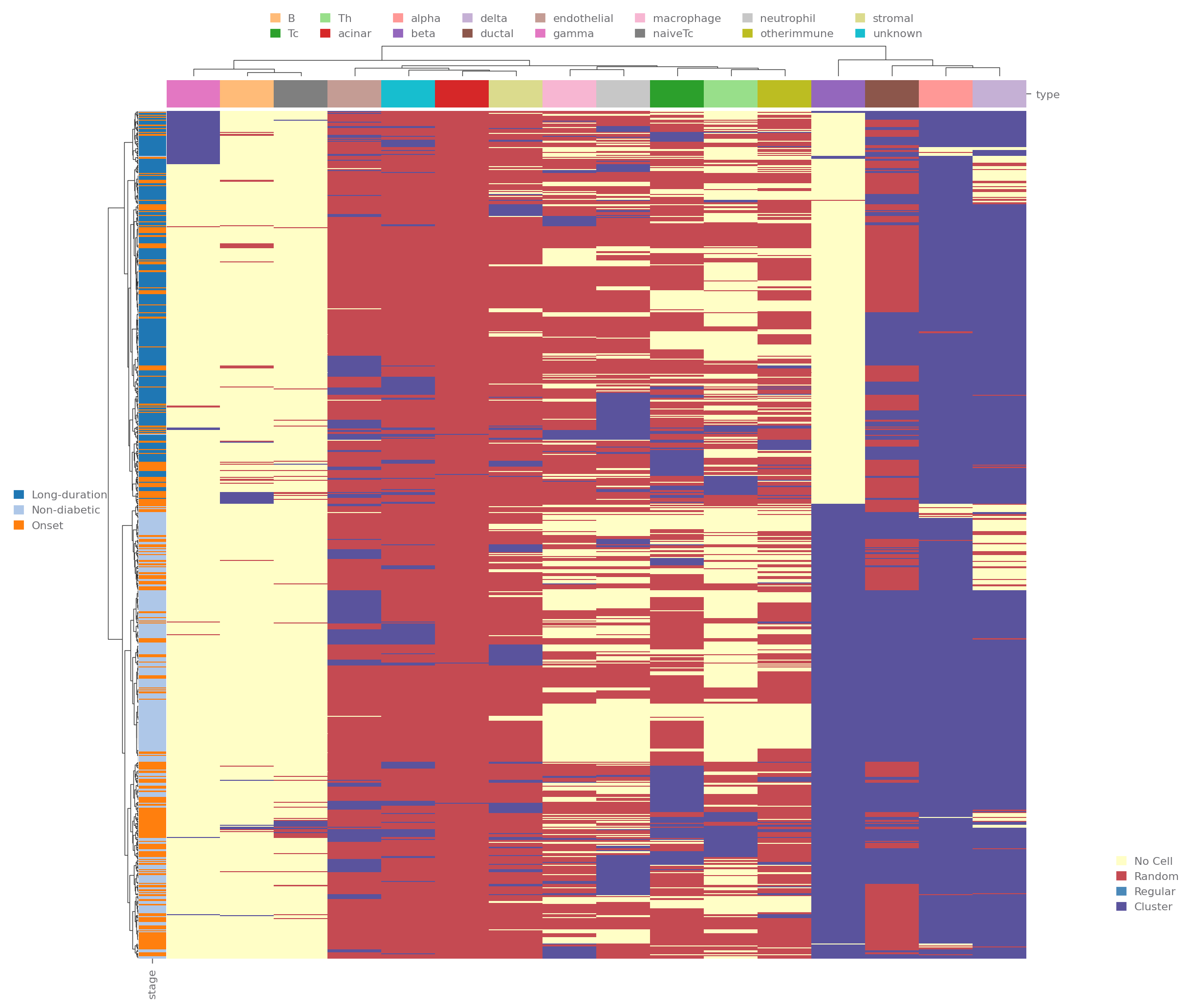
[7]:
Spatial Heterogeneity¶
Used to evaluate the tissue heterogeneity
Three methods are provided:
Shannon entropy (No spatial information)
Leibovici entropy (Spatial entropy)
Altieri entropy (Spatial entropy)
The number of three species are the same in three figures, You can see that shannon entropy remain the same. Only spatial entropy is changing.

[8]:
st.spatial_heterogeneity(data)
⏳ Spatial Heterogeneity
🛠 Method: Leibovici entropy
(pid=2801) Error in atexit._run_exitfuncs:
(pid=2801) Traceback (most recent call last):
(pid=2801) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2801) self._writeout_input_cache(conn)
(pid=2801) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2801) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2801) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145521209344 and this is thread id 4697800192.
(pid=2790) Error in atexit._run_exitfuncs:
(pid=2790) Traceback (most recent call last):
(pid=2790) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2790) self._writeout_input_cache(conn)
(pid=2790) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2790) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2790) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145471897600 and this is thread id 4530720256.
(pid=2800) Error in atexit._run_exitfuncs:
(pid=2800) Traceback (most recent call last):
(pid=2800) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2800) self._writeout_input_cache(conn)
(pid=2800) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2800) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2800) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145565806592 and this is thread id 4557991424.
(pid=2791) Error in atexit._run_exitfuncs:
(pid=2791) Traceback (most recent call last):
(pid=2791) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2791) self._writeout_input_cache(conn)
(pid=2791) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2791) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2791) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145411981312 and this is thread id 4718685696.
(pid=2796) Error in atexit._run_exitfuncs:
(pid=2796) Traceback (most recent call last):
(pid=2796) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2796) self._writeout_input_cache(conn)
(pid=2796) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2796) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2796) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145353105408 and this is thread id 4552564224.
(pid=2787) Error in atexit._run_exitfuncs:
(pid=2787) Traceback (most recent call last):
(pid=2787) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2787) self._writeout_input_cache(conn)
(pid=2787) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2787) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2787) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145352572928 and this is thread id 4407406080.
(pid=2789) Error in atexit._run_exitfuncs:
(pid=2789) Traceback (most recent call last):
(pid=2789) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 780, in writeout_cache
(pid=2789) self._writeout_input_cache(conn)
(pid=2789) File "/Users/milk/anaconda3/envs/spatialtis/lib/python3.8/site-packages/IPython/core/history.py", line 763, in _writeout_input_cache
(pid=2789) conn.execute("INSERT INTO history VALUES (?, ?, ?, ?)",
(pid=2789) sqlite3.ProgrammingError: SQLite objects created in a thread can only be used in that same thread. The object was created in thread id 123145462366208 and this is thread id 4593577472.
Calculating heterogeneity ██████████ 100% 00:00|00:01
📦 Added to AnnData, uns: 'spatial_heterogeneity'
⏱ 44s234ms
[8]:
[9]:
sp.spatial_heterogeneity(data, ['stage'], size=(4, 4),
group_order=group_order,
title="Tissue heterogeneity in diabetic stages",
yaxis_title="Heterogeneity")
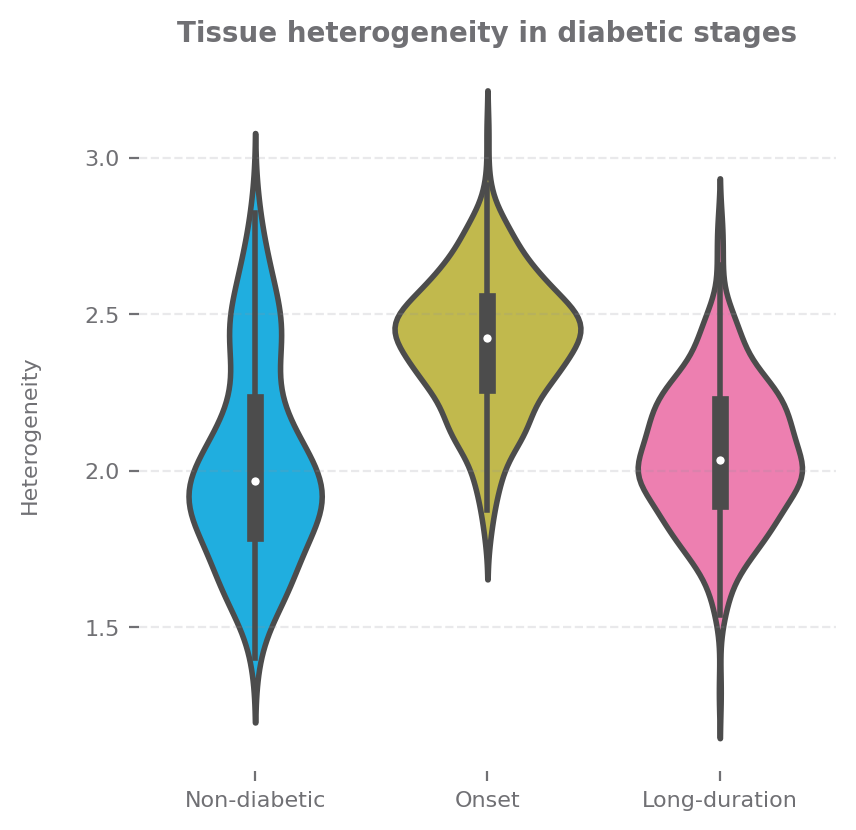
[9]:
If we look at cell components, we could see that the cell type are more divese in Onset stage
[10]:
st.cell_components(data)
⏳ Cell Components
📦 Added to AnnData, uns: 'cell_components'
⏱ 524ms
[10]:
[11]:
cells = ['Tc', 'Th', 'naiveTc', 'macrophage', 'neutrophil', 'otherimmune', 'alpha', 'beta', 'gamma', 'delta']
sp.cell_components(data, ['stage'], agg="mean", percentage=False, selected_types=cells, group_order=group_order, size=(3,3.5),
title="Immune cells in Diabetes Stage")
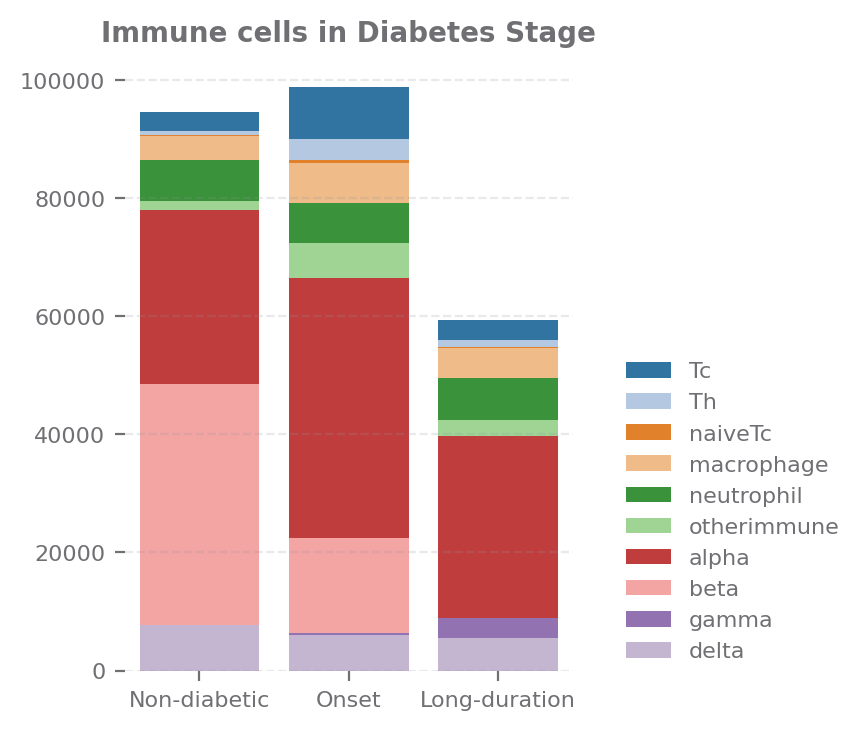
[11]:
Hotspot detection¶
Spatial distribution tells you if a cell is clustered, hotspot detection tells you which cells are clustered.
[12]:
st.hotspot(data, selected_types=islets_cells, grid_size=50, search_level=2)
⏳ Hotspot Analysis
Hotspot analysis ██████████ 100% 00:00|00:00
📦 Added to AnnData, obs: 'hotspot'
⏱ 6s396ms
[12]:
[13]:
st.hotspot(data, selected_types=['alpha'], grid_size=15, search_level=2)
⏳ Hotspot Analysis
Hotspot analysis ██████████ 100% 00:00|00:00
📦 Added to AnnData, obs: 'hotspot'
⏱ 6s955ms
[13]:
[14]:
ROI = {"stage": "Onset", "image": "A01"}
sp.cell_map(data, ROI, cell_type_key="hotspot", selected_types=['cold', 'hot'], palette="Paired")
sp.cell_map(data, ROI, selected_types=islets_cells)
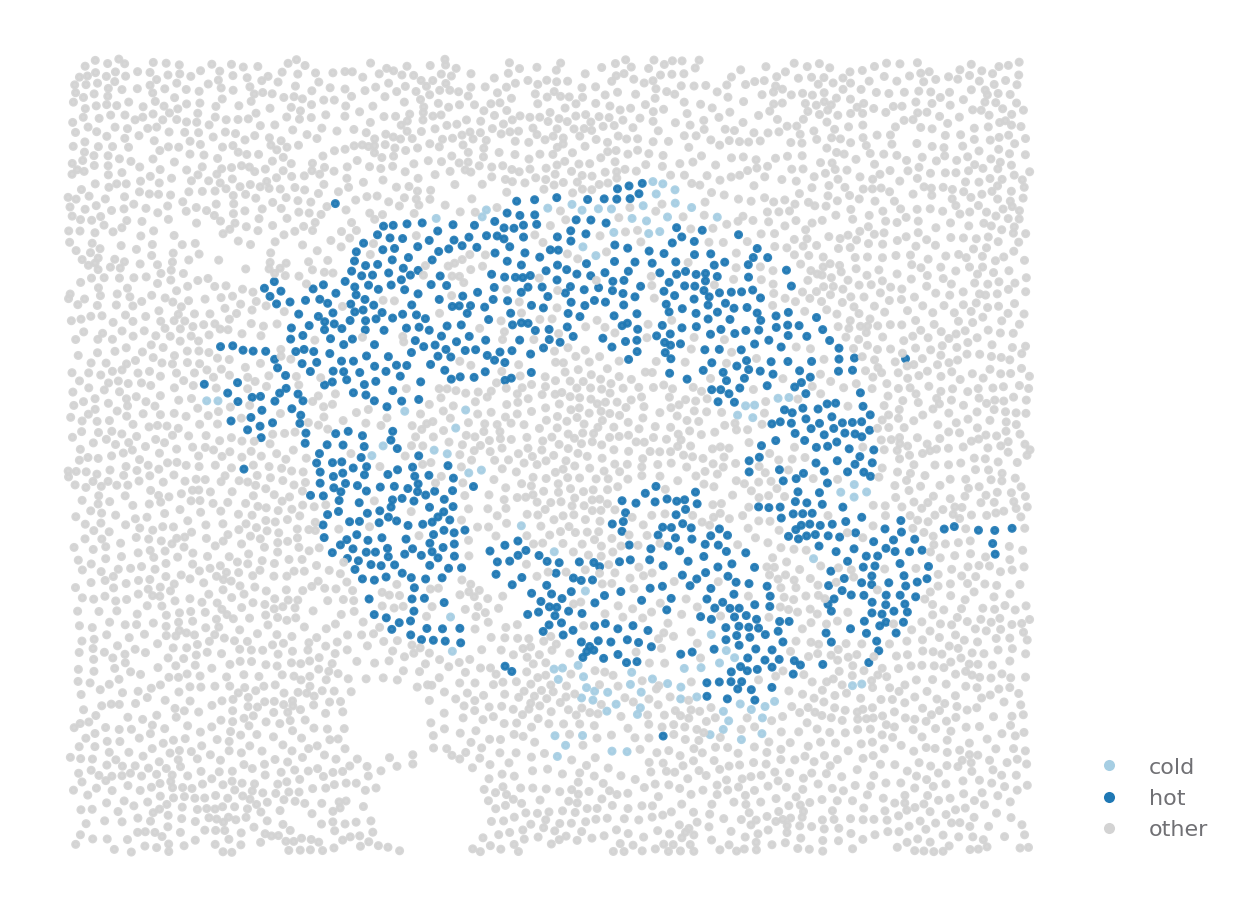
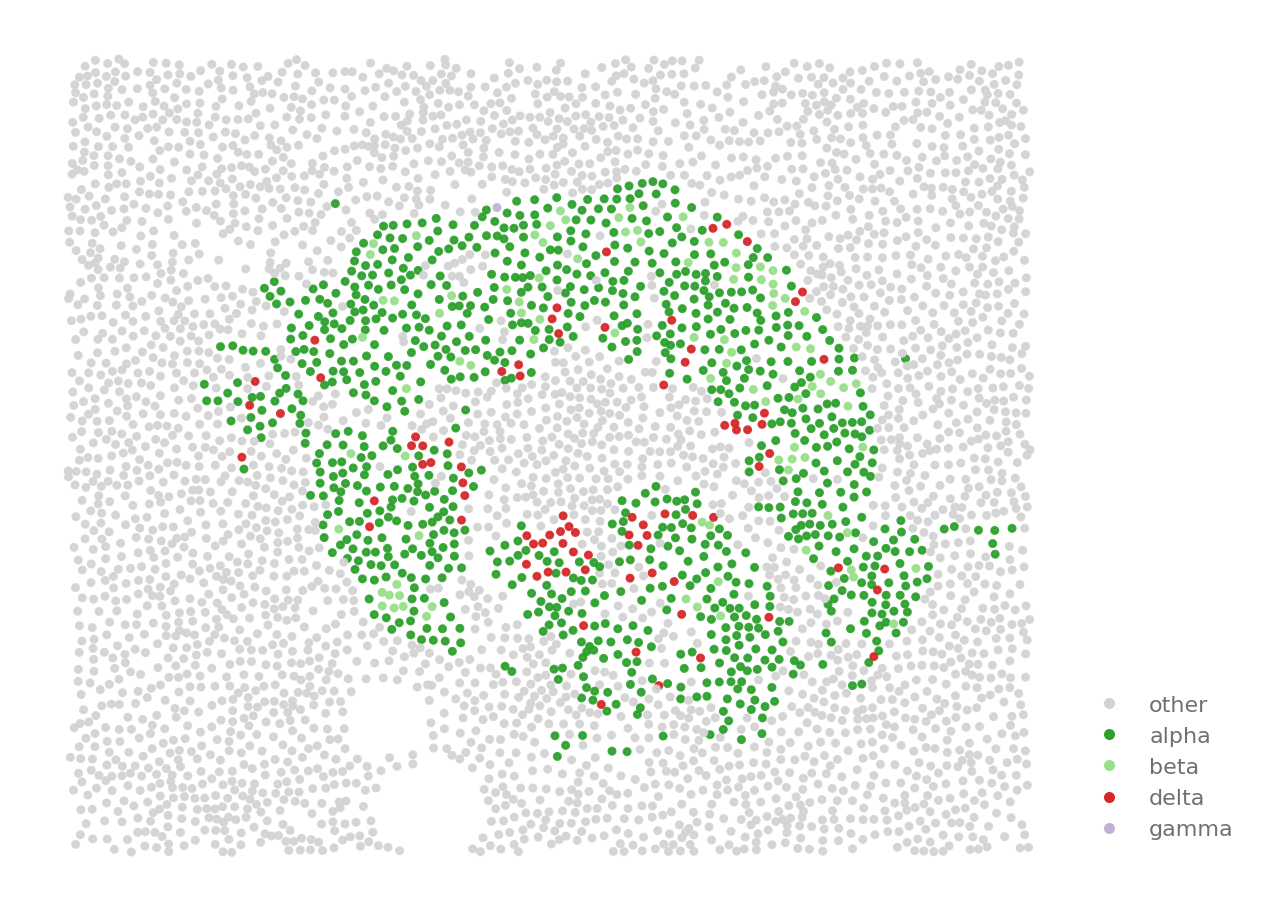
[14]:
Find cell neighbors¶
Every analysis below will rely on this step. This is a critical step.
Two methods are provided:
KD-tree: If your cells are points
R-tree: If your cells are polygons
This method is implemented in Rust, it’s much faster even that scipy’s implementation of KD-tree in C
This dataset only contains points information.
[15]:
st.find_neighbors(data, expand=8)
⏳ Find Cell Neighbors
🛠 Method: KD-tree
Find neighbors ██████████ 100% 00:00|00:14
📦 Added to AnnData, obs: 'cell_neighbors'
📦 Added to AnnData, obs: 'cell_neighbors_count'
⏱ 17s907ms
[15]:
Neighborhood Analysis¶
To profile the cell-cell interaction
This method is also implemented in Rust, see how fast it is.
Three viusalizations:
Dot matrix plot
Heatmap
Graph (static / interactive)
[16]:
st.neighborhood_analysis(data)
⏳ Neighborhood Analysis
🛠 Method: pseudo p-value
Neighborhood analysis ██████████ 100% 00:00|00:28
📦 Added to AnnData, uns: 'neighborhood_analysis'
⏱ 30s493ms
[16]:
[17]:
sp.neighborhood_analysis(data)
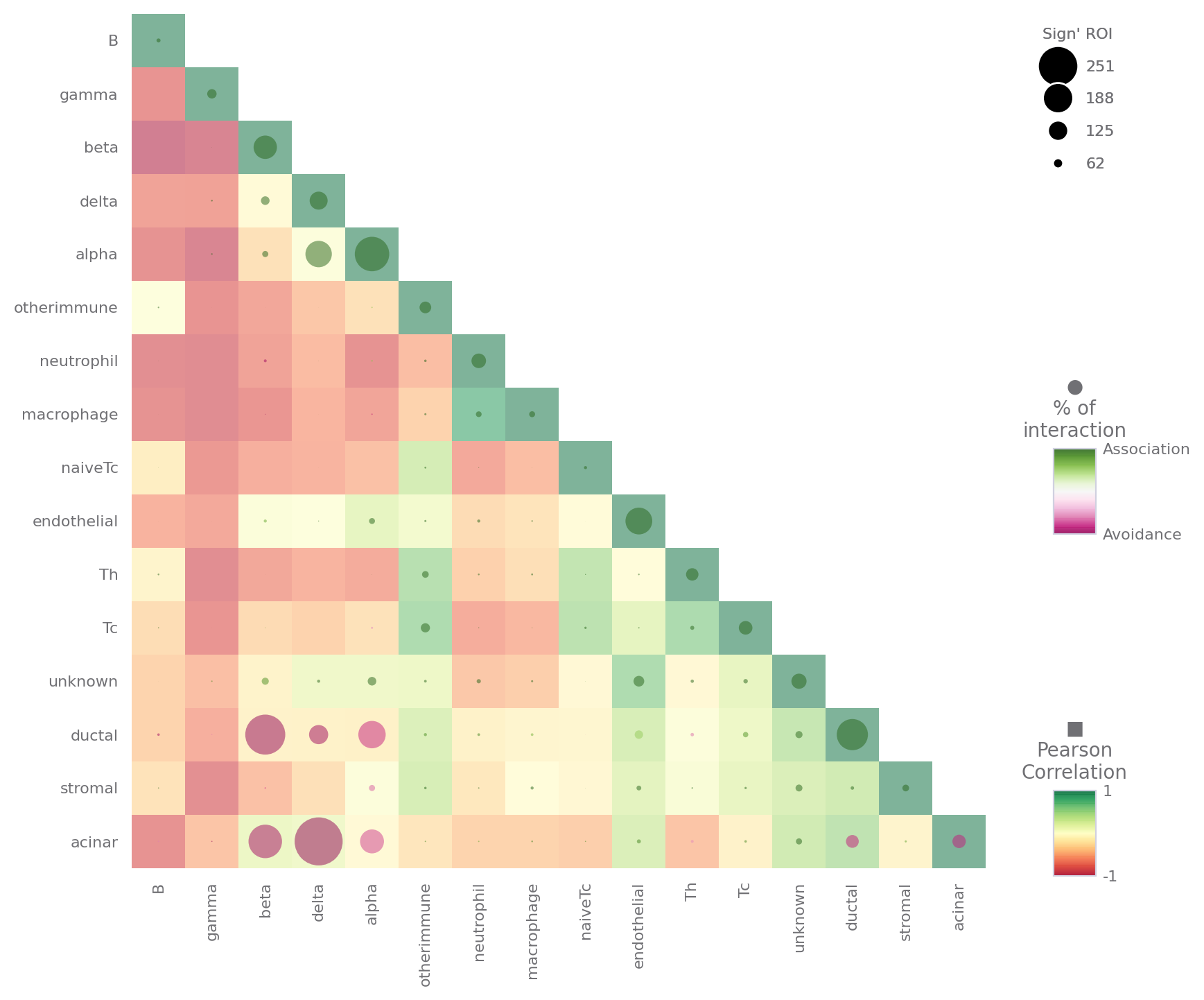
[17]:
[18]:
cell_type_order = ['alpha', 'beta', 'delta', 'gamma', 'Tc', 'naiveTc', 'Th', 'B', 'macrophage', 'neutrophil',
'otherimmune', 'acinar', 'ductal', 'endothelial', 'stromal', 'unknown']
sp.neighborhood_analysis(data, selected_types=cell_type_order, use="heatmap",
clustermap_kwargs=dict(row_cluster=False, col_cluster=False, method="ward"))
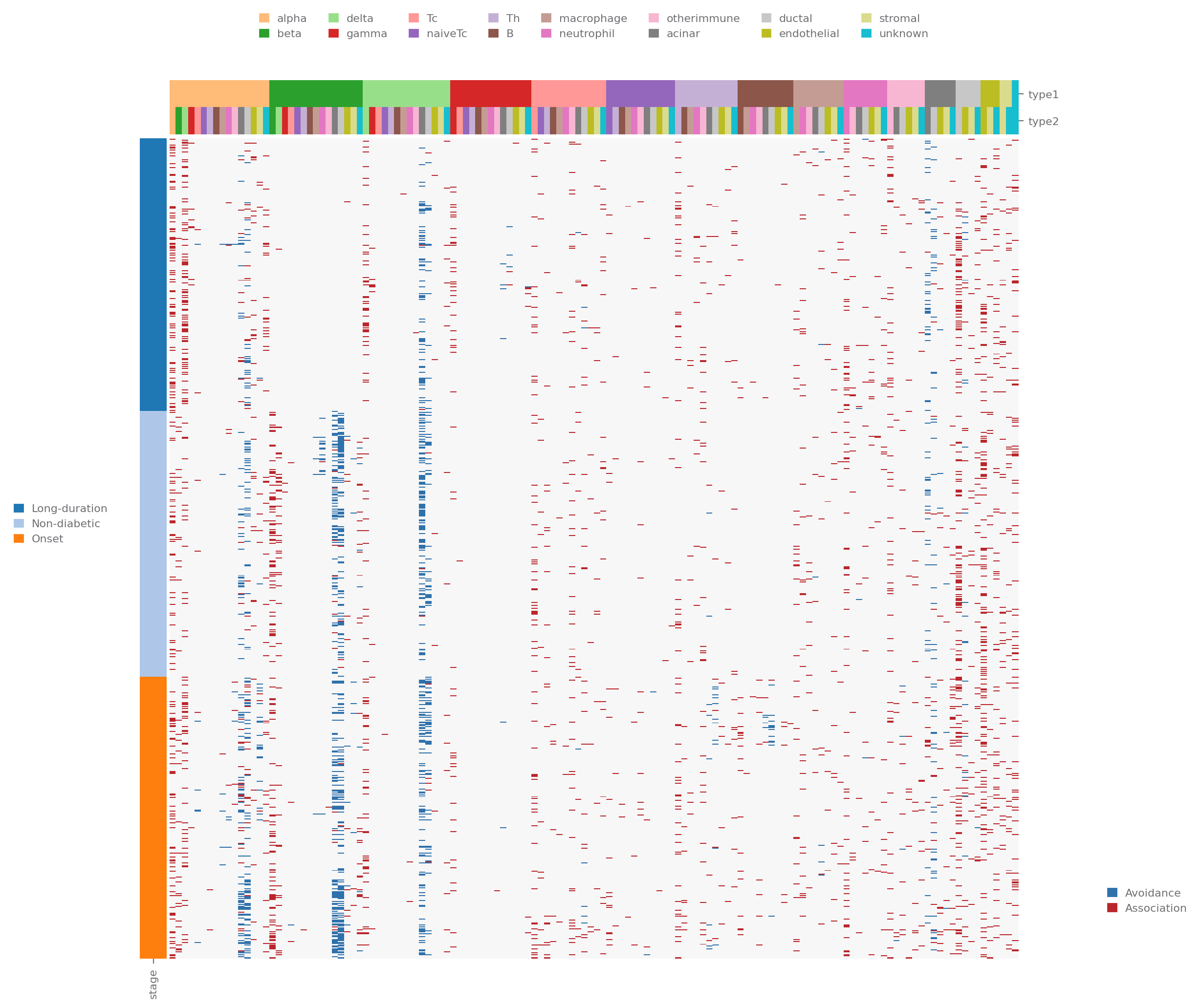
[18]: